2024
Andrussow, I., Sun, H., Martius, G., Kuchenbecker, K. J.
Demonstration: Minsight - A Soft Vision-Based Tactile Sensor for Robotic Fingertips
Hands-on demonstration presented at the Conference on Robot Learning (CoRL), Munich, Germany, November 2024 (misc) Accepted
Cao, C. G. L., Javot, B., Bhattarai, S., Bierig, K., Oreshnikov, I., Volchkov, V. V.
Fiber-Optic Shape Sensing Using Neural Networks Operating on Multispecklegrams
IEEE Sensors Journal, 24(17):27532-27540, September 2024 (article)
Bizeul, A., Schölkopf, B., Allen, C.
A Probabilistic Model behind Self-Supervised Learning
Transactions on Machine Learning Research, September 2024 (article) To be published
Chen*, W., Horwood*, J., Heo, J., Hernández-Lobato, J. M.
Leveraging Task Structures for Improved Identifiability in Neural Network Representations
Transactions on Machine Learning Research, August 2024, *equal contribution (article)
Kladny, K., Kügelgen, J. V., Schölkopf, B., Muehlebach, M.
Deep Backtracking Counterfactuals for Causally Compliant Explanations
Transactions on Machine Learning Research, July 2024 (article)
Schölkopf, B.
Grundfragen der künstlichen Intelligenz
astronomie - Das Magazin, 42, May 2024 (article)
Zabel, S., Hennig, P., Nieselt, K.
VIPurPCA: Visualizing and Propagating Uncertainty in Principal Component Analysis
IEEE Transactions on Visualization and Computer Graphics, 30(4):2011-2022, April 2024 (article)
Rahaman, N., Weiss, M., Wüthrich, M., Bengio, Y., Li, E., Pal, C., Schölkopf, B.
Language Models Can Reduce Asymmetry in Information Markets
arXiv:2403.14443, March 2024, Published as: Redesigning Information Markets in the Era of Language Models, Conference on Language Modeling (COLM) (techreport)
Mancini, M., Naeem, M. F., Xian, Y., Akata, Z.
Learning Graph Embeddings for Open World Compositional Zero-Shot Learning
IEEE Transactions on Pattern Analysis and Machine Intelligence, 46(3):1545-1560, IEEE, New York, NY, March 2024 (article)
Visonà, G., Bouzigon, E., Demenais, F., Schweikert, G.
Network propagation for GWAS analysis: a practical guide to leveraging molecular networks for disease gene discovery
Briefings in Bioinformatics, 25(2), February 2024 (article)
Pals, M., Macke, J. H., Barak, O.
Trained recurrent neural networks develop phase-locked limit cycles in a working memory task
PLOS Computational Biology, 20(2), February 2024 (article)
Peisen, F., Gerken, A., Dahm, I., Nikolaou, K., Eigentler, T., Amaral, T., Moltz, J. H., Othman, A. E., Gatidis, S.
Pre-treatment 18F-FDG-PET/CT parameters as biomarkers for progression free survival, best overall response and overall survival in metastatic melanoma patients undergoing first-line immunotherapy
PLOS ONE, 19(1), January 2024 (article)
Villar, S., Hogg, D. W., Yao, W., Kevrekidis, G. A., Schölkopf, B.
Towards fully covariant machine learning
Transactions on Machine Learning Research, January 2024 (article)
Bonse, M. J., Gebhard, T. D., Dannert, F. A., Absil, O., Cantalloube, F., Christiaens, V., Cugno, G., Garvin, E. O., Hayoz, J., Kasper, M., Matthews, E., Schölkopf, B., Quanz, S. P.
Use the 4S (Signal-Safe Speckle Subtraction): Explainable Machine Learning reveals the Giant Exoplanet AF Lep b in High-Contrast Imaging Data from 2011
2024 (misc) Submitted
Gebhard, T. D., Angerhausen, D., Konrad, B. S., Alei, E., Quanz, S. P., Schölkopf, B.
Parameterizing pressure-temperature profiles of exoplanet atmospheres with neural networks
Astronomy & Astrophysics, 681, 2024 (article)
Tsirtsis, S., Tabibian, B., Khajehnejad, M., Singla, A., Schölkopf, B., Gomez-Rodriguez, M.
Optimal Decision Making Under Strategic Behavior
Management Science, 2024, Published Online (article) In press
Rajendran, G., Buchholz, S., Aragam, B., Schölkopf, B., Ravikumar, P.
Learning Interpretable Concepts: Unifying Causal Representation Learning and Foundation Models
2024 (misc)
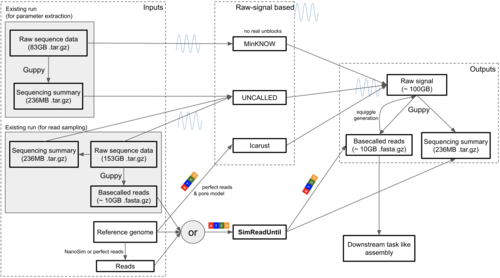
Mordig, M., Ratsch, G., Kahles, A.
SimReadUntil for benchmarking selective sequencing algorithms on ONT devices
Bioinformatics, 40, 2024 (article)
2023
Visonà, G., Spiller, L. M., Hahn, S., Hattingen, E., Vogl, T. J., Schweikert, G., Bankov, K., Demes, M., Reis, H., Wild, P., Zeiner, P. S., Acker, F., Sebastian, M., Wenger, K. J.
Machine-Learning-Aided Prediction of Brain Metastases Development in Non-Small-Cell Lung Cancers
Clinical lung cancer, 24(8):e311-e322, December 2023 (article)
Visonà, G., Duroux, D., Miranda, L., Sükei, E., Li, Y., Borgwardt, K., Oliver, C.
Multimodal learning in clinical proteomics: enhancing antimicrobial resistance prediction models with chemical information
Bioinformatics, 39(12), December 2023 (article)
Laumann, F., von Kügelgen, J., Park, J., Schölkopf, B., Barahona, M.
Kernel-Based Independence Tests for Causal Structure Learning on Functional Data
Entropy, 25(12), November 2023 (article)
Kerber, B., Hepp, T., Küstner, T., Gatidis, S.
Deep learning-based age estimation from clinical Computed Tomography image data of the thorax and abdomen in the adult population
PLOS ONE, 18(11), November 2023 (article)
Lei, A., Schölkopf, B., Posner, I.
Variational Causal Dynamics: Discovering Modular World Models from Interventions
Transactions on Machine Learning Research, November 2023 (article)
Cao, X., Liu, W., Tsang, I. W.
Data-Efficient Learning via Minimizing Hyperspherical Energy
IEEE transactions on pattern analysis and machine intelligence, 45(11):13422-13437, November 2023 (article)
Feuerecker, B., Heimer, M. M., Geyer, T., Fabritius, M. P., Gu, S., Schachtner, B., Beyer, L., Ricke, J., Gatidis, S., Ingrisch, M., Cyran, C. C.
Artificial Intelligence in Oncological Hybrid Imaging
Nuklearmedizin, 62(5):296-305, October 2023 (article)
Hayoz, J., Cugno, G., Quanz, S. P., Patapis, P., Alei, E., Bonse, M. J., Dannert, F. A., Garvin, E. O., Gebhard, T. D., Konrad, B. S., Sartori, L. F.
CROCODILE - Incorporating medium-resolution spectroscopy of close-in directly imaged exoplanets into atmospheric retrievals via cross-correlation
Astronomy & Astrophysics, 678, October 2023 (article)
Hupkes, D., Giulianelli, M., Dankers, V., Artetxe, M., Elazar, Y., Pimentel, T., Christodoulopoulos, C., Lasri, K., Saphra, N., Sinclair, A., Ulmer, D., Schottmann, F., Batsuren, K., Sun, K., Sinha, K., Khalatbari, L., Ryskina, M., Frieske, R., Cotterell, R., Jin, Z.
A taxonomy and review of generalization research in NLP
Nature Machine Intelligence, 5(10):1161-1174, October 2023 (article)
Peisen, F., Gerken, A., Dahm, I., Nikolaou, K., Eigentler, T., Amaral, T., Moltz, J. H., Othman, A. E., Gatidis, S., Dondi, F.
Can Whole-Body Baseline CT Radiomics Add Information to the Prediction of Best Response, Progression-Free Survival, and Overall Survival of Stage IV Melanoma Patients Receiving First-Line Targeted Therapy: A Retrospective Register Study
Diagnostics (Basel), 13(20), October 2023 (article)
Ke*, N. R., Bilaniuk*, O., Goyal, A., Bauer, S., Larochelle, H., Schölkopf, B., Mozer, M. C., Pal, C., Bengio, Y.
Neural Causal Structure Discovery from Interventions
Transactions on Machine Learning Research, September 2023, *equal contribution (article)
Malinverno, L., Barros, V., Ghisoni, F., Visonà, G., Kern, R., Nickel, P. J., Ventura, B. E., Šimić, I., Stryeck, S., Manni, F., Ferri, C., Jean-Quartier, C., Genga, L., Schweikert, G., Lovrić, M., Rosen-Zvi, M.
A historical perspective of biomedical explainable AI research
Patterns, 4(9), September 2023 (article)
Buchholz, S.
Some Remarks on Identifiability of Independent Component Analysis in Restricted Function Classes
Transactions on Machine Learning Research, September 2023 (article)
Boelts, J., Harth, P., Gao, R., Udvary, D., Yáñez, F., Baum, D., Hege, H., Oberlaender, M., Macke, J. H.
Simulation-based inference for efficient identification of generative models in computational connectomics
PLOS Computational Biology, 19(9):1-28, September 2023 (article)
Hawkins-Hooker, A., Visonà, G., Narendra, T., Rojas-Carulla, M., Schölkopf, B., Schweikert, G.
Getting personal with epigenetics: towards individual-specific epigenomic imputation with machine learning
Nature Communications, 14(1), August 2023 (article)
Vaughan, S. R., Gebhard, T. D., Bott, K., Casewell, S. L., Cowan, N. B., Doelman, D. S., Kenworthy, M., Mazoyer, J., Millar-Blanchaer, M. A., Trees, V. J. H., Stam, D. M., Absil, O., Altinier, L., Baudoz, P., Belikov, R., Bidot, A., Birkby, J. L., Bonse, M. J., Brandl, B., Carlotti, A., Choquet, E., van Dam, D., Desai, N., Fogarty, K., Fowler, J., van Gorkom, K., Gutierrez, Y., Guyon, O., Haffert, S. Y., Herscovici-Schiller, O., Hours, A., Juanola-Parramon, R., Kleisioti, E., König, L., van Kooten, M., Krasteva, M., Laginja, I., Landman, R., Leboulleux, L., Mouillet, D., N’Diaye, M., Por, E. H., Pueyo, L., Snik, F.
Chasing rainbows and ocean glints: Inner working angle constraints for the Habitable Worlds Observatory
Monthly Notices of the Royal Astronomical Society, 524(4):5477-5485, August 2023 (article)
Bonse, M. J., Garvin, E. O., Gebhard, T. D., Dannert, F. A., Cantalloube, F., Cugno, G., Absil, O., Hayoz, J., Milli, J., Kasper, M., Quanz, S. P.
Comparing Apples with Apples: Robust Detection Limits for Exoplanet High-contrast Imaging in the Presence of Non-Gaussian Noise
The American Astronomical Society, 166(2), July 2023 (article)
Wellnitz, D., Kekić, A., Heiss, J., Gertz, M., Weidemüller, M., Spitz, A.
A network approach to atomic spectra
Journal of Physics: Complexity, 4(3), July 2023 (article)
Ortiz-Jimenez*, G., de Jorge*, P., Sanyal, A., Bibi, A., Dokania, P. K., Frossard, P., Rogez, G., Torr, P.
Catastrophic overfitting can be induced with discriminative non-robust features
Transactions on Machine Learning Research , July 2023, *equal contribution (article)
Wang, Q., Sanchez, F. R., McCarthy, R., Bulens, D. C., McGuinness, K., O’Connor, N., Wüthrich, M., Widmaier, F., Bauer, S., Redmond, S. J.
Dexterous robotic manipulation using deep reinforcement learning and knowledge transfer for complex sparse reward-based tasks
Expert Systems, 40(6), July 2023 (article)
Kekić, A., Dehning, J., Gresele, L., von Kügelgen, J., Priesemann, V., Schölkopf, B.
Evaluating vaccine allocation strategies using simulation-assisted causal modeling
Patterns, 4(6), June 2023 (article)
Katiyar, P., Schwenck, J., Frauenfeld, L., Divine, M. R., Agrawal, V., Kohlhofer, U., Gatidis, S., Kontermann, R., Königsrainer, A., Quintanilla-Martinez, L., la Fougère, C., Schölkopf, B., Pichler, B. J., Disselhorst, J. A.
Quantification of intratumoural heterogeneity in mice and patients via machine-learning models trained on PET–MRI data
Nature Biomedical Engineering, 7(8):1014-1027, June 2023 (article)
Badirli, S., Picard, C. J., Mohler, G., Richert, F., Akata, Z., Dundar, M.
Classifying the unknown: Insect identification with deep hierarchical Bayesian learning
Methods in Ecology and Evolution, 14(6):1515-1530, June 2023 (article)
Mineeva*, O., Danciu*, D., Schölkopf, B., Ley, R. E., Rätsch, G., Youngblut, N. D.
ResMiCo: Increasing the quality of metagenome-assembled genomes with deep learning
PLOS Computational Biology, 19(5), Public Library of Science, San Francisco, CA, May 2023, *equal contribution (article)
Gatidis, S., Kart, T., Fischer, M., Winzeck, S., Glocker, B., Bai, W., Bülow, R., Emmel, C., Friedrich, L., Kauczor, H., Keil, T., Kröncke, T., Mayer, P., Niendorf, T., Peters, A., Pischon, T., Schaarschmidt, B., Schmidt, B., Schulze, M., Umutle, L., Völzke, H., Küstner, T., Bamberg, F., Schölkopf, B., Rueckert, D.
Better Together: Data Harmonization and Cross-StudAnalysis of Abdominal MRI Data From UK Biobank and the German National Cohort
Investigative Radiology, 58(5):346-354, May 2023 (article)
Kanagawa, H., Jitkrittum, W., Mackey, L., Fukumizu, K., Gretton, A.
A Kernel Stein Test for Comparing Latent Variable Models
Journal of the Royal Statistical Society Series B: Statistical Methodology, 85(3):986-1011, May 2023 (article)
Blum, D., Hepp, T., Belov, V., Goya-Maldonado, R., la Fougère, C., Reimold, M.
Estimating uncertainty in read-out patterns: Application to controls-based denoising and voxel-based morphometry patterns in neurodegenerative and neuropsychiatric diseases
Human Brain Mapping, 44(7):2802-2814, May 2023 (article)
Kim, J., Singh, S., Vales, C., Keebler, E., Fisher, A. V., Thiessen, E. D.
Staying and Returning dynamics of young children’s attention
Developmental Science, 26(6), May 2023 (article)
Safavi, S., Panagiotaropoulos, T. I., Kapoor, V., Ramirez-Villegas, J. F., Logothetis, N., Besserve, M.
Uncovering the Organization of Neural Circuits with Generalized Phase Locking Analysis
PLOS Computational Biology, 19(4):45, Public Library of Science, April 2023 (article)
Reizinger, P., Sharma, Y., Bethge, M., Schölkopf, B., Huszár, F., Brendel, W.
Jacobian-based Causal Discovery with Nonlinear ICA
Transactions on Machine Learning Research, April 2023 (article)
Schreiber*, J., Boix*, C., Lee, J. W., Li, H., Guan, Y., Chang, C., Chang, J., Hawkins-Hooker, A., Schölkopf, B., Schweikert, G., Carulla, M. R., Canakoglu, A., Guzzo, F., Nanni, L., Masseroli, M., Carman, M. J., Pinoli, P., Hong, C., Yip, K. Y., Spence, J. P., Batra, S. S., Song, Y. S., Mahony, S., Zhang, Z., Tan, W., Shen, Y., Sun, Y., Shi, M., Adrian, J., Sandstrom, R., Farrell, N., Halow, J., Lee, K., Jiang, L., Yang, X., Epstein, C., Strattan, J. S., Bernstein, B., Snyder, M., Kellis, M., Stafford, W., Kundaje, A., ENCODE Imputation Challenge Participants,
The ENCODE Imputation Challenge: a critical assessment of methods for cross-cell type imputation of epigenomic profiles
Genome Biology, 24, April 2023, *co‑first authors (article)